Ranking co-occurrences in the scientific literature using the Log of the Product of Frequency (LPF).
The LPF is not sensitive to literature volume. Use of the LPF for ranking co-occurrences helps investigators explore pathway, disease, and intervention concurrence in the literature with greater efficiency and accuracy.
By using the LPF, Literature Lab™ avoids lenses associated with terms or genes that have large publication volume. The LPF is a measure of the intensity of the co-occurrences as measured in the literature. This technique may prevent an association from “disappearing” in cases where a term that may not have significant publication volume has the bulk of its appearances in specific relationships.
The LPF is used with all co-occurrence presentations on genes and biological and biochemical concepts in the Literature Lab™ database.
The LPF is computed as follows:
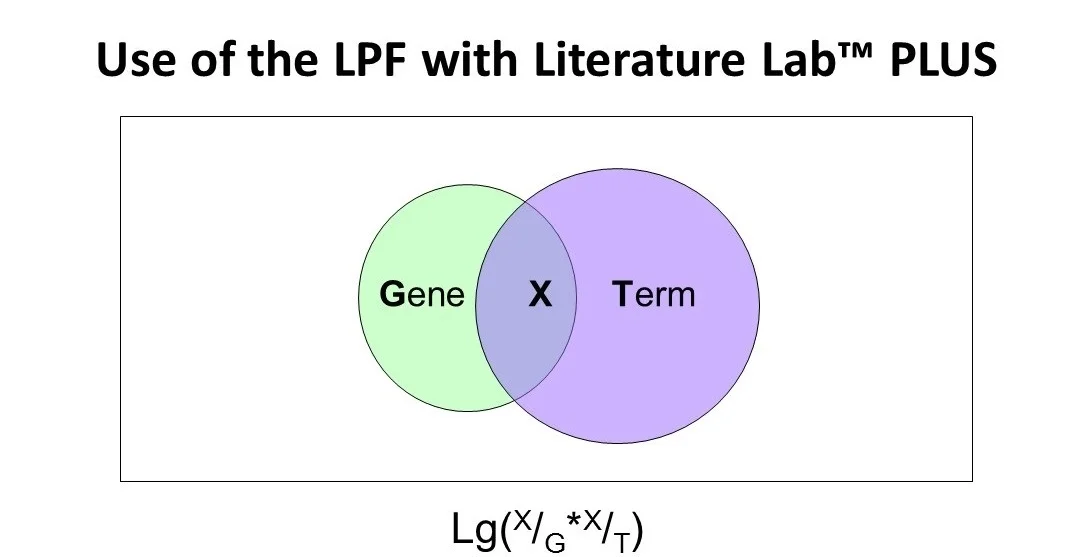
For each Term in the Literature Lab™ database (see first image above):
Term 1 represents the number of abstracts that contain one term (in a collection of 86,000 terms)
Term 2 represents the number of abstracts that contain another term
X is the number of abstracts mentioning both terms
P1 = Term 1/X - the proportion of abstracts mentioning first term
P2 = Term 2/X - the proportion of abstracts mentioning the second term
P = P1 * P2 - the product of the proportions
The logarithm of P is computed to compensate for the attenuation bias from multiplying two fractions.
Co-occurrences are evaluated in Literature Lab™ PLUS using the same technique. The LPFs are computed for each gene in a gene set and each of the 86,000 terms, providing an analysis of relative intensity of associations in the literature for the gene set. When multiple genes are mentioned with a term the LPFs are added, resulting in an LPF for the gene set with the term. The Literature Lab™ PLUS analysis thus is an analysis on the gene set as a set. The LPFs of the submitted gene set are compared to the same computations applied to 1,000 random gene sets to determine significance. Z-score and P-values are presented, and significant associations are highlighted. The literature Lab PLUS gene list analyzer wraps up the analysis by computing a clustering analysis on the significantly associated functions with the genes.
The LPF is always a negative number (except when the circles in the Venn diagram are congruent) and the larger the absolute magnitude of the LPF, the weaker the association relative to other gene/term or term/term co-occurrences in a dimension, e.g.: pathways or diseases, as shown in the examples on the Literature Lab™ page.
1. Yanhui Hu, Lisa M. Hines, Haifeng Weng, Dongmei Zuo, Miguel Rivera, Andrea Richardson, and Joshua LaBaer. Analysis of Genomic and Proteomic Data Using Advanced Literature Mining. Journal of Proteome Research, 2003, 2, 405-412 PMID: 12938930